What Do Supervised and Unsupervised Actually Mean in Machine Learning?
In machine learning there are many umbrellas that refer to different processes or set ups for the “learning” to take place. Today’s discussion will surround two commonly used umbrella terms, supervised and unsupervised learning. In this case the process or set up being referred to actually lays within the raw data itself.
When setting up a machine learning project, there are a few considerations that shape the nature of the project. The first is the type of data available, either categorical or continuous. The second is whether or not a ground truth is available. It should be noted that not all numerical data is continuous. The commonly used Likert scale testing, answering a question with a response of 1 to 5 with one being the worst and five being the best, is actually categorical data, because there is no float type, like 1.5034, answer possible. This 1.5034 response would be sorted into either the 1 or the 2 category, making the data type categorical. Whereas in the continuous datatype, any number along a number line or curve is a valid data point. An example of categorical data in the field of oil and gas, specifically in drilling, is lithology. An example of continuous data would be something like mechanical specific energy (mse) or weight on bit (wob). Whether the type of data is categorical or continuous is only one consideration. Additionally, there needs to be a decision about whether or not there is some sort of “ground truth” associated with the data.
This “ground truth” functions like an answer key in that you, as the human, already know the correct answer before asking the question of an artificial intelligence or constructing the machine learning model. One version of this is having labeled data available for categorical data, such as having the lithology determined for every depth when trying to classify lithology. Another version would be having something like a wire line log available to compare a predicted wire line log against. Having this ground truth available for use, means that a supervised machine learning model can be selected. Otherwise when this type of “ground truth” data is not available, the options for a machine learning project are restricted to only unsupervised machine learning techniques. If you have this answer key, you can choose either a supervised or unsupervised approach, or sometimes both, to see which will yield better results, and you can actually use both to have them compete against each other in a reinforcement learning scenario. Basically, if you start with some knowledge that is reliable in the real world, a “ground truth”, then you have more options in creating your artificial world using machine learning.
With this in mind, data quality is a huge consideration when embarking on a project. If there is partial labeled data available, is it enough to construct a supervised learning model? Or would an unsupervised approach be better with wider access to include the unlabeled data? One way to assist in making these decisions is to consider the problem being addressed. Has any work been done before to connect any of the variables or features at play? What is the nature of this relationship? Is it predictable? In the case of oil and gas, which uses geomechanical properties that have been studied for over a century, often times this answer lies within the physical relationships being studied. Meaning that sometimes, there’s a way to solve for a missing label algebraically. This takes some time in writing the code to execute that formula, but also it means working closely with someone like a petroleum engineer to make sure that all of those geomechanical relationships are being referenced and used appropriately. There’s also the additional step of associating that newly solved for label with the dataset being worked with. But this would take a dataset previously only being able to be used in unsupervised learning to having more options under a labeled dataset, now under the supervised learning umbrella.
Something that also plays into the decision about using a supervised or unsupervised approach is the testing or validation of that machine learning model. In supervised learning the testing is really easy. You just compare the results from the model to that “ground truth” or answer key to determine accuracy. There are some considerations to be had with under- and over-fitting, but at the end of the day, calculating that accuracy is just like your teacher grading your test in school. For some business questions, an accuracy of 70% is good enough. For other questions, often those with high risk associated with failure, an accuracy of 95% or better might be required. It should be noted that an accuracy of 100% definitely means that the model is overfit. This means that when introduced to a new dataset, the model will likely fail because it is different than the training dataset. This under- and over-fitting problem is a drawback to using a supervised approach, you have to hit that sweet spot of accuracy.
All of these considerations, while they still apply in unsupervised learning situations, are much more difficult to assess in an unsupervised environment. In this case, measuring some inherent statistical properties in model outputs and making sure that they are in line with other similar problems is a good route to take in determining model performance. In the case of unsupervised learning, the conversation surrounding accuracy is null and void because there’s no answer key. So the conversation must switch frame to answering the question of is this output realistic? Does is make sense? Is it reproducible and to what scale? Often times in unsupervised scenarios a multitude of models are generated to assess performance in comparison to each other. This is much like taste testing some food after selecting a different quality of ingredients for each preparation of the same dish. Some will behave similarly, others will be awful, and a few might be really good. There’s not really a way to know beforehand though that the store bought brand actually does better than the branded product, not until you try it. Each type of unsupervised machine learning model has its own metric by which it can be evaluated, and this is how comparison of or model performance is discussed in an unsupervised scenario. A discussion before beginning any modeling is in order to determine if that metric will do enough to assess performance to an acceptable standard.
With all of this in mind, conversations surrounding what data is available and how it needs to be evaluated will help in the decision making process of not only whether or not a machine learning option is a viable path forward, but also what type of machine learning algorithm to use, in making the decision about how to address a business question or need in industry. This conversation should also include what types of resources need to be dedicated to the problem. While all types of machine learning takes tuning, unsupervised learning models do present more challenge, and often require manpower and more computational time to generate the variety of models for comparison.
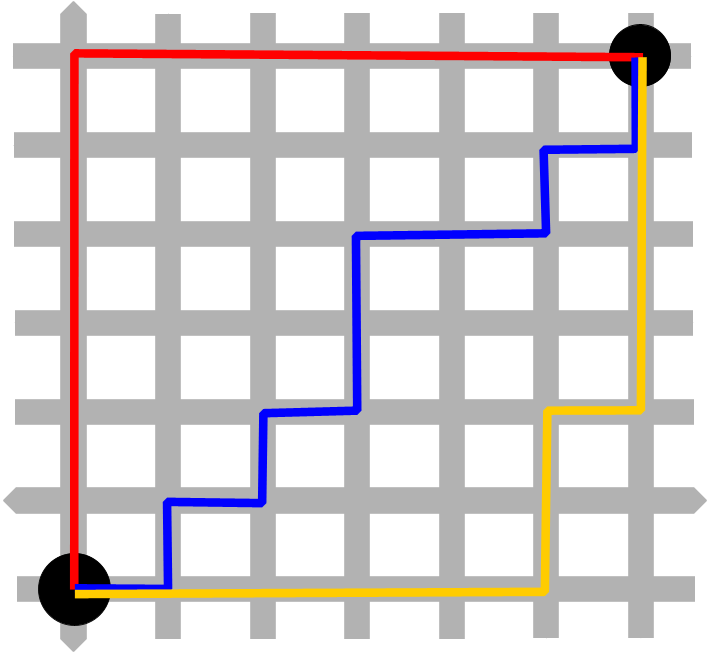 Figure 1. Manhattan distance grid pattern.
Figure 1. Manhattan distance grid pattern.
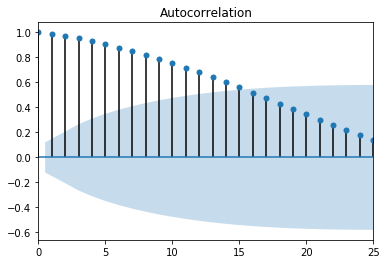 Figure 1. ACF random walk
Figure 1. ACF random walk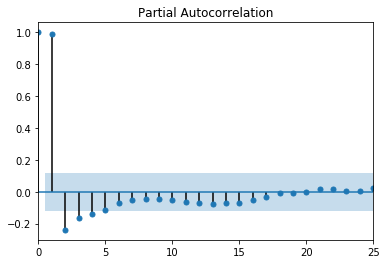 Figure 2. PACF random walk
Figure 2. PACF random walk Figure 1. The Scientific Method as an Ongoing Process
Figure 1. The Scientific Method as an Ongoing Process Figure 2. The OSEMN Data Science Method
Figure 2. The OSEMN Data Science Method